
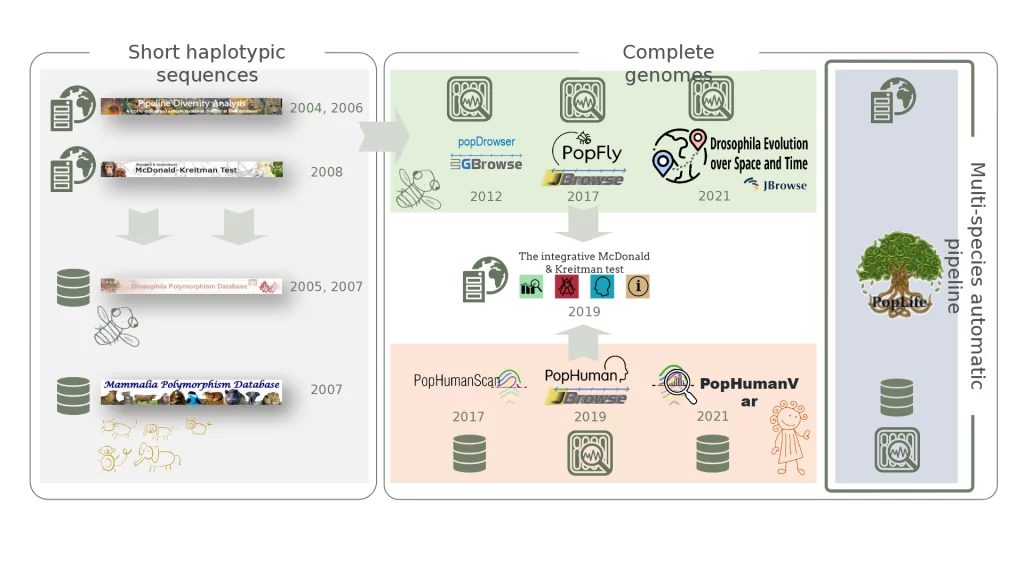
Multi-species automatic pipeline

PopLife 
PopLife is a free online resource for researchers studying population variation in genomes. It uses both traditional and AI-based methods to extract knowledge from genomic data. It applies to various fields like evolutionary genomics, functional genomics, conservation genetics, and more. PopLife aims to become an international reference for population genomics
Complete genomes

PopDrowser 
‘Population Drosophila Browser’ (PopDrowser), is a new genome browser specially designed for the automatic analysis and representation of genetic variation across the D. melanogaster genome sequence.
Publications:

PopFly 
PopFly is a population genomics-oriented genome browser, based on JBrowse software, that contains a complete inventory of population genomic parameters estimated from DGN data.
Publications:

Drosophilia Evolution over Space and Time 
Drosophila Evolution over Space and Time (DEST) is coupled with sampling and environmental metadata. A web-based genome browser and web portal provide easy access to the SNP data set.
Publications:

The integrative McDonald & Kreitman Test 
iMKT (acronym for integrative McDonald and Kreitman test), is a novel web-based service performing four distinct MKT types. It allows the detection and estimation of four different selection regimes −adaptive, neutral, strongly deleterious and weakly deleterious− acting on any genomic sequence.
Publications:

PopHumanScan 
PopHumanScan, an online catalog that compiles and annotates all candidate regions under selection to facilitate their validation and thoroughly analysis.
Publications:

PopHuman 
PopHuman is a population genomics-oriented genome browser, based on JBrowse, that contains a complete inventory of nucleotide diversity metrics, linkage disequilibrium, recombination rates and neutrality tests in sliding windows along the human genome estimated from the 1000 Genomes Project data using the PopGenome software.
Publications:

PopHumanVar 
PopHumanVar is an interactive application designed to facilitate the exploration and thorough analysis of candidate genomic regions, generating useful summary reports of the variants that are putatively causal of recent selective sweeps.
Publications:
Short haplotypic sequences

Pipeline Diversity Analisys 
PDA, “Pipeline Diversity Analysis”, is a collection of programs and modules mainly written in Perl that automatically can search for polymorphic sequences in a large database, and estimate their genetic diversity.
Publications:

Standard & Generated McDonald-Kreitman Test 
The McDonald and Kreitman test (MKT) is a widely used method to detect natural selection at the molecular level. We introduce a new website, the standard and generalized MKT website, which expands the test’s capabilities to include other regions and loci. The website has three interfaces: standard MKT for analyzing coding region sites, advanced MKT for comparing closely linked coding or noncoding regions, and multi-locus MKT for analyzing multiple loci. The website has demonstrated the correlation between selection efficiency and effective population size in Drosophila, and it has been used to estimate selection in DPDB. This timely resource is expected to be widely adopted by researchers and contribute to the catalog of adaptive evolution cases.
Publications:

Drosophila Polymorphism Database
DPDB, ‘Drosophila Polymorphism Database’, is a web site that provides a daily updated repository of all well-annotated polymorphic sequences in the Drosophila genus. It allows the search for any polymorphic set according to different parameter values of nucleotide diversity, linkage disequilibrium and codon bias.
Publications:

Mammalia Polymorphism Database
MamPol, ‘Mammalia Polymorphism Database’, is a website containing all the well-annotated polymorphic sequences available in GenBank for the Mammalia class grouped by name of organism and gene.
Publications: